Molecules 1.1 submitted for review
This might be jumping the gun a bit, but I wanted to let you know that a new version of Molecules, 1.1, has been submitted for review for the iTunes App Store. Even though it will take a bit of time to be approved, I thought you might be interested in a preview of some of the new features in this version.
This version adds two new visualization modes, as well as integrated keyword searching of the Protein Data Bank.
The new visualization modes are cylinders and spacefilling. The cylindrical representation shows only the bonds, and gives those bonds a color based on their amino acid or nucleotide residue. The colors have been drawn from Jmol's color scheme. The spacefilling mode draws atoms according to their van der Waals radius. Unfortunately, this mode doesn't provide the best visual experience, because it exposes the icosahedrons I use for representing spheres. I will be adjusting the rendering of this mode to smooth out the jagged edges.
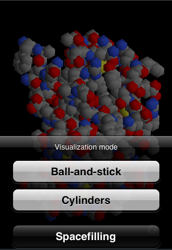
To switch to these new rendering modes, double-tap on the 3-D model. This will bring up a sheet where you can pick a new visualization mode.
The user interface has also been cleaned up slightly by the conversion of titles and other entries in the PDB records to title case. The ALL-CAPS was hurting my eyes. Thanks go to Marshall Elfstrand for his Objective-C version of John Gruber's title case Perl script.
A common complaint with Molecules is how hard it is to get new structures into the program. To improve this, I've replaced the old system of entering PDB codes by hand with the ability to perform a direct keyword search of the Protein Data Bank. Now, if you tap on "Download new molecules", you can enter keywords in the following search box and titles and PDB codes of matching entries will be displayed in the table below. Tapping on a title will bring you to a screen that lets you view the molecule's entry on the Protein Data Bank web page or download the structure to the iPhone (or iPod Touch).
This keyword searching mechanism is not perfect, because it relies on a relatively new web services API at the Protein Data Bank. Currently, results to the keyword search are returned in order of entry to the data bank, not in order of relevance.  Also, I'm capping results at about 50 right now to save on bandwidth when communicating over the cell networks. I display titles for the search results because that is the only data field that can be retrieved given the search results using the current API. However, it sounds like there is significant interest at the Protein Data Bank to expand these services in the near future, so hopefully this search functionality will become even more powerful and user friendly.
Also, I'm capping results at about 50 right now to save on bandwidth when communicating over the cell networks. I display titles for the search results because that is the only data field that can be retrieved given the search results using the current API. However, it sounds like there is significant interest at the Protein Data Bank to expand these services in the near future, so hopefully this search functionality will become even more powerful and user friendly.
I'd like to personally thank Dr. Rachel Kramer Green and Wayne Townsend-Merino of the Protein Data Bank for their help with the integration of the web services in Molecules.
Unfortunately, I wanted to focus on adding these new features for this version, so no significant improvements have been made to the performance of the application. This will be my next priority. As discussed here, I plan to re-implement the molecule data model as a SQLite database and cache the render buffers to disk, hopefully providing nearly instantaneous load times and very fast render speeds. I also will work to fix the memory issues affecting very large molecules and try to optimize the OpenGL rendering speeds.
As usual, I'll post when this goes live in the App Store.